Information of ICE
| ICEberg ID | 193_IME |
|---|---|
| ICE Name | NBU1 |
| ICEO ID | - |
| Organism | Bacteroides uniformis |
| Size | 10276 bp |
| GC Content (%) | 40.24 |
| Insertion site | 3′ of a tRNALeu gene |
| Function | - |
| Species that ICE can be transferred to | Bacteroides spp.; Escherichia coli |
| Nucleotide Sequence | AF238307 (complete IME sequence in this GenBank file) |
| Coordinates | 1..10276 |
| Putative oriT region | coordinates: 8213..8426; oriTDB id: 200010 CGCAAGGAATACAGCAGAGACCGATACATCCGTGACGCTTCACAGATTTACAGCGGATGC AAGGACTTGAACGAATACTTACAGAAACAGGTTGAAAGAAAAAGGCAAGTCCAATCCGTC AAAGGGATGAGCAGCCAGTCACCGAAAAAGAAAAACGGCTTTCGGTTATAGCCCACTATA ACTACCTCCGCTTTCGTAGTTGTGGGCTCTCC |
| Putative relaxase | coordinates: 8592..9995; Family: - |
Function tags R Antibiotic resistance V Virulence factor M Metal resistance D Defense system G Degradation S Symbiosis
The graph information of NBU1 components from AF238307
Complete gene list of NBU1 from AF238307
| # | Gene | Coordinates [+/-], size (bp) | Product (GenBank annotation) | Reannotation | Rea |
|---|
The Interaction Network among ICE/IME/CIME
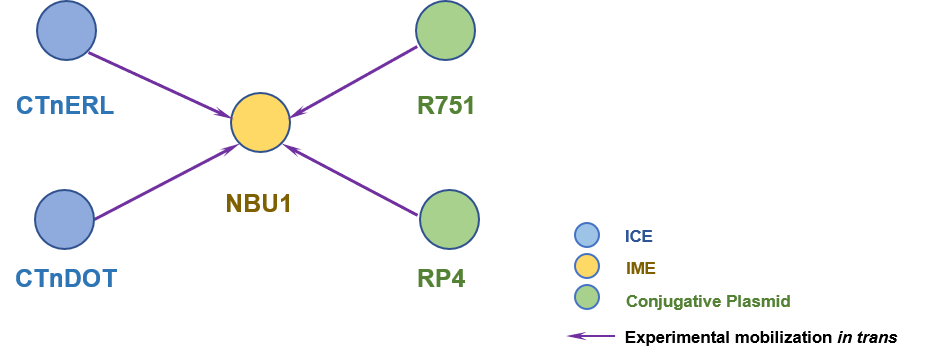
Detailed Informatioin of the Interaction Network
| # | ICE | Inter_Ele [Type] | Methods | Donors | Recipients | Exper_Ref |
|---|---|---|---|---|---|---|
| 1 | NBU1 | CTnERL [ICE] | | Bacteroides uniformis | Bacteroides thetaiotaomicron 5482; Escherichia coli | 8407835;7608064 |
| 2 | NBU1 | CTnDOT [ICE] | | Bacteroides uniformis | Bacteroides thetaiotaomicron 5482 | 8407835 |
| 3 | NBU1 | RP4 [IncP plasmid] | | E. coli DH5αMCR | E. coli HB101 | 8407836 |
| 4 | NBU1 | R751 [IncP plasmid] | | Escherichia coli | Escherichia coli; Bacteroides thetaiotaomicron | 8407835; 8407836 |
| (1) Bass KA, Hecht DW. (2002) Isolation and characterization of cLV25, a Bacteroides fragilis chromosomal transfer factor resembling multiple Bacteroides sp. mobilizable transposons. J Bacteriol. 2002 Apr;184(7):1895-904. [PubMed:11889096] |
| (2) Stevens AM, Shoemaker NB, Salyers AA. (1990) The region of a Bacteroides conjugal chromosomal tetracycline resistance element which is responsible for production of plasmidlike forms from unlinked chromosomal DNA might also be involved in transfer of the element. J Bacteriol. 1990 Aug;172(8):4271-9. [PubMed:2165473] |
| (3) Shoemaker NB, Salyers AA. (1988) Tetracycline-dependent appearance of plasmidlike forms in Bacteroides uniformis 0061 mediated by conjugal Bacteroides tetracycline resistance elements. J Bacteriol. 1988 Apr;170(4):1651-7. [PubMed:2832373] |
| (4) Li LY, Shoemaker NB, Wang GR, Cole SP, Hashimoto MK, Wang J, Salyers AA. (1995) The mobilization regions of two integrated Bacteroides elements, NBU1 and NBU2, have only a single mobilization protein and may be on a cassette. J Bacteriol. 1995 Jul;177(14):3940-5. [PubMed:7608064] |
| (5) Shoemaker NB, Li LY, Salyers AA. (1994) An unusual type of cointegrate formation between a Bacteroides plasmid and the excised circular form of an integrated element (NBU1). Plasmid. 1994 Nov;32(3):312-7. [PubMed:7899516] |
| (6) Shoemaker NB, Wang GR, Stevens AM, Salyers AA. (1993) Excision, transfer, and integration of NBU1, a mobilizable site-selective insertion element. J Bacteriol. 1993 Oct;175(20):6578-87. [PubMed:8407835] |
| (7) Li LY, Shoemaker NB, Salyers AA. (1993) Characterization of the mobilization region of a Bacteroides insertion element (NBU1) that is excised and transferred by Bacteroides conjugative transposons. J Bacteriol. 1993 Oct;175(20):6588-98. [PubMed:8407836] |
| (8) Shoemaker NB, Wang GR, Salyers AA. (1996) The Bacteroides mobilizable insertion element, NBU1, integrates into the 3' end of a Leu-tRNA gene and has an integrase that is a member of the lambda integrase family. J Bacteriol. 1996 Jun;178(12):3594-600. [PubMed:8655559] |
| (9) Shoemaker NB, Wang GR, Salyers AA. (1996) NBU1, a mobilizable site-specific integrated element from Bacteroides spp., can integrate nonspecifically in Escherichia coli. J Bacteriol. 1996 Jun;178(12):3601-7. [PubMed:8655560] |
| (10) Cooper AJ, Kalinowski AP, Shoemaker NB, Salyers AA. (1997) Construction and characterization of a Bacteroides thetaiotaomicron recA mutant: transfer of Bacteroides integrated conjugative elements is RecA independent. J Bacteriol. 1997 Oct;179(20):6221-7. [PubMed:9335266] |
| (11) Shoemaker NB, Wang GR, Salyers AA. (2000) Multiple gene products and sequences required for excision of the mobilizable integrated Bacteroides element NBU1. J Bacteriol. 2000 Feb;182(4):928-36. [PubMed:10648516] |
| (12) Wang J, Shoemaker NB, Wang GR, Salyers AA. (2000) Characterization of a Bacteroides mobilizable transposon, NBU2, which carries a functional lincomycin resistance gene. J Bacteriol. 2000 Jun;182(12):3559-71. [PubMed:10852890] |
| (13) Wang J, Wang GR, Shoemaker NB, Salyers AA. (2001) Production of two proteins encoded by the Bacteroides mobilizable transposon NBU1 correlates with time-dependent accumulation of the excised NBu1 circular form. J Bacteriol. 2001 Nov;183(21):6335-43. [PubMed:11591678] |
| (14) Rajeev L, Salyers AA, Gardner JF. (2006) Characterization of the integrase of NBU1, a Bacteroides mobilizable transposon. Mol Microbiol. 2006 Aug;61(4):978-90. [PubMed:16859497] |
| (15) Rajeev L, Segall A, Gardner J. (2007) The bacteroides NBU1 integrase performs a homology-independent strand exchange to form a holliday junction intermediate. J Biol Chem. 2007 Oct 26;282(43):31228-37. [PubMed:17766246] |
| (16) Song B, Wang GR, Shoemaker NB, Salyers AA. (2009) An unexpected effect of tetracycline concentration: growth phase-associated excision of the Bacteroides mobilizable transposon NBU1. J Bacteriol. 2009 Feb;191(3):1078-82. [PubMed:18952794] |
| (17) Rajeev L, Malanowska K, Gardner JF. (2009) Challenging a paradigm: the role of DNA homology in tyrosine recombinase reactions. Microbiol Mol Biol Rev. 2009 Jun;73(2):300-9. [PubMed:19487729] |
| (18) Bellanger X, Payot S, Leblond-Bourget N, Guédon G. (2014) Conjugative and mobilizable genomic islands in bacteria: evolution and diversity. FEMS Microbiol Rev. 2014 Jul;38(4):720-60. [PubMed:24372381] |
| (19) Guédon G, Libante V, Coluzzi C, Payot S, Leblond-Bourget N. (2017) The Obscure World of Integrative and Mobilizable Elements, Highly Widespread Elements that Pirate Bacterial Conjugative Systems. Genes (Basel). 2017 Nov 22;8(11):337. [PubMed:29165361] |