
| 1. How to use T4SEfinder Web Server |
| 1.1. Input protein sequence(s) in the FASTA format |
 |
First, the user should prepare the protein sequence(s) under study in the FASTA format. The protein sequences can either be typed directly into the text area, or can be uploaded from a file using the button.
Only the FASTA Format is acceptable to the T4SEfinder server. The lines starting with '>' are treated as the identifiers of the following sequences. All sequences have to consist of 20 common amino acids specified in a single letter code. T4SEfinder will also check the empty sequence and the uniqueness of sequence identifiers. In addition, the input webpage also provides an example. Please click the "Show an Example" to display an example input for T4SEfinder. You can also click "example.fasta" to download an example file. |
| 1.2. Select the prediction algorithm |
 |
You can choose the prediction algorithm among TAPEBert_MLP, PSSM_CNN and HybridBiLSTM (the maximum numbers of input protein sequences are 6000, 250 and 250, respectively). After submition, T4SEfinder will produce a Job ID and a web link to the results.
|
| 1.3. Retrieve the completed results |
 |
You can track your job status and check the completed results by Job ID in the retrieve page.
|
| 2. Interpretation of the prediction result |
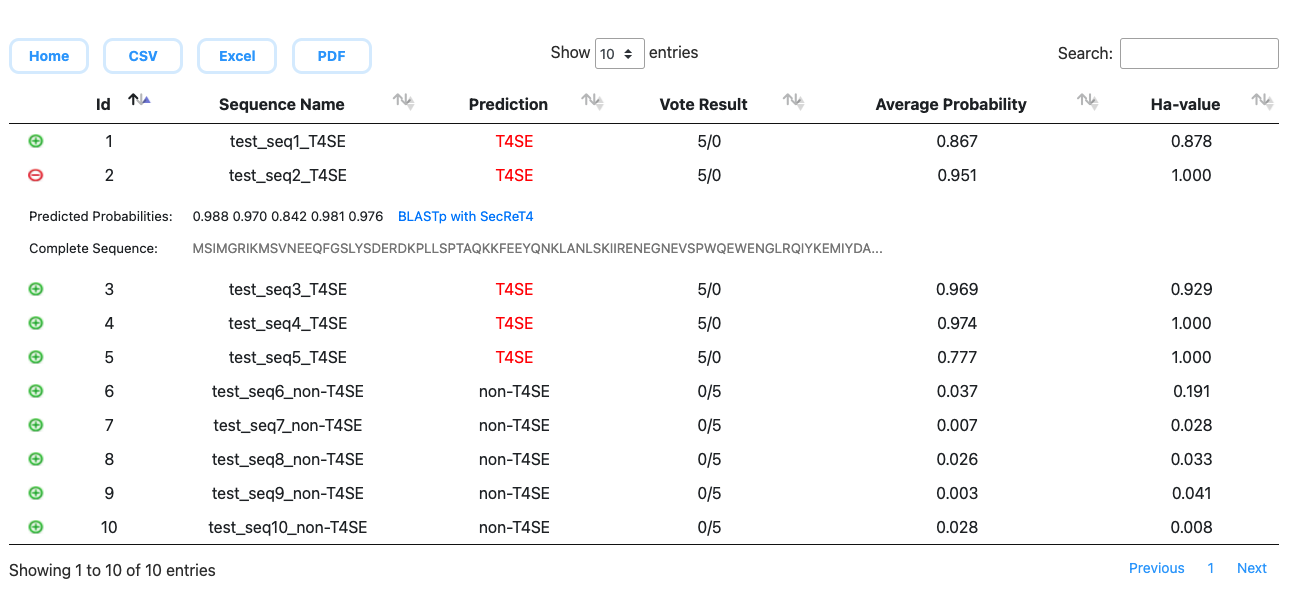 |
Prediction: Final predicted label (T4SE or non-T4SE) for the query protein according to the vote result.
Vote Result: Voting result by predictors derived from cross-validation. (numbers to support T4SE or non-T4SE) Average Probability: Average predicted probability as a T4SE by predictors derived from cross-validation. Ha-value: A sequence similarity measure between the query protein and the experimentally verified ones. All predicted probabilities and complete query sequence are demonstrated in the extended columns. You can download the prediction result in the format of CSV, Excel, PDF files. You can also search the similar structure or target proteins of putative T4SEs in the SecReT4 database. |