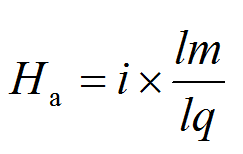To examine the degree of sequence similarities at an amino acid level between each query protein and the T4SEfinder-collected
T4SEs, the NCBI BLASTp-derived
Ha-value was employed. For each query, the
Ha-value was calculated as follows:
where
i was the level of BLASTp identities of the region with the highest Bit score expressed as a frequency of between
0 and 1,
lm the length of the highest scoring matching sequence (including gaps) and
lq the
query length. If there were no matching sequences with a BLASTp
E-value < 0.01, the
Ha-value assigned to that
query sequence was defined as zero. Therefore
Ha-value belonged to the set,
Ha ∈[0,1]. Here, a strict
Ha-value cut-off
≥ 0.42 was used to determine the significant sequence similarities.