Information of ICE
| ICEberg ID | 238_IME |
|---|---|
| ICE Name | tISCpe8 |
| ICEO ID | - |
| Organism | Clostridium perfringens |
| Size | 1964 bp |
| GC Content (%) | 26.63 |
| Insertion site | AT-rich regions |
| Function | R Lincosamide |
| Species that ICE can be transferred to | - |
| Nucleotide Sequence | FJ589781 (complete IME sequence in this GenBank file) |
| Coordinates | 1..1964 |
| Putative oriT region | - |
| Putative relaxase | - |
Function tags R Antibiotic resistance V Virulence factor M Metal resistance D Defense system G Degradation S Symbiosis
The graph information of tISCpe8 components from FJ589781
Complete gene list of tISCpe8 from FJ589781
| # | Gene | Coordinates [+/-], size (bp) | Product (GenBank annotation) | Reannotation | Rea |
|---|
The Interaction Network among ICE/IME/CIME
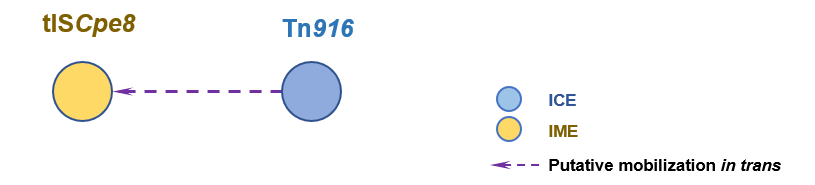
Detailed Informatioin of the Interaction Network
| # | ICE | Inter_Ele [Type] | Methods | Donors | Recipients | Exper_Ref |
|---|---|---|---|---|---|---|
| 1 | tISCpe8 | Tn916 [ICE] | | Clostridium perfringens JIR4225 | Clostridium perfringens JIR4394 | 19684139 |
| (1) Lyras D, Adams V, Ballard SA, Teng WL, Howarth PM, Crellin PK, Bannam TL, Songer JG, Rood JI. (2009) tISCpe8, an IS1595-family lincomycin resistance element located on a conjugative plasmid in Clostridium perfringens. J Bacteriol. 2009 Oct;191(20):6345-51. [PubMed:19684139] |
| (2) Bellanger X, Payot S, Leblond-Bourget N, Guédon G. (2014) Conjugative and mobilizable genomic islands in bacteria: evolution and diversity. FEMS Microbiol Rev. 2014 Jul;38(4):720-60. [PubMed:24372381] |
| (3) Guédon G, Libante V, Coluzzi C, Payot S, Leblond-Bourget N. (2017) The Obscure World of Integrative and Mobilizable Elements, Highly Widespread Elements that Pirate Bacterial Conjugative Systems. Genes (Basel). 2017 Nov 22;8(11):337. [PubMed:29165361] |